
David Twesigomwe — PhD Graduate, University of the Witwatersrand
August 8, 2022
Unisa adds academic muscle to African genome research
September 20, 2022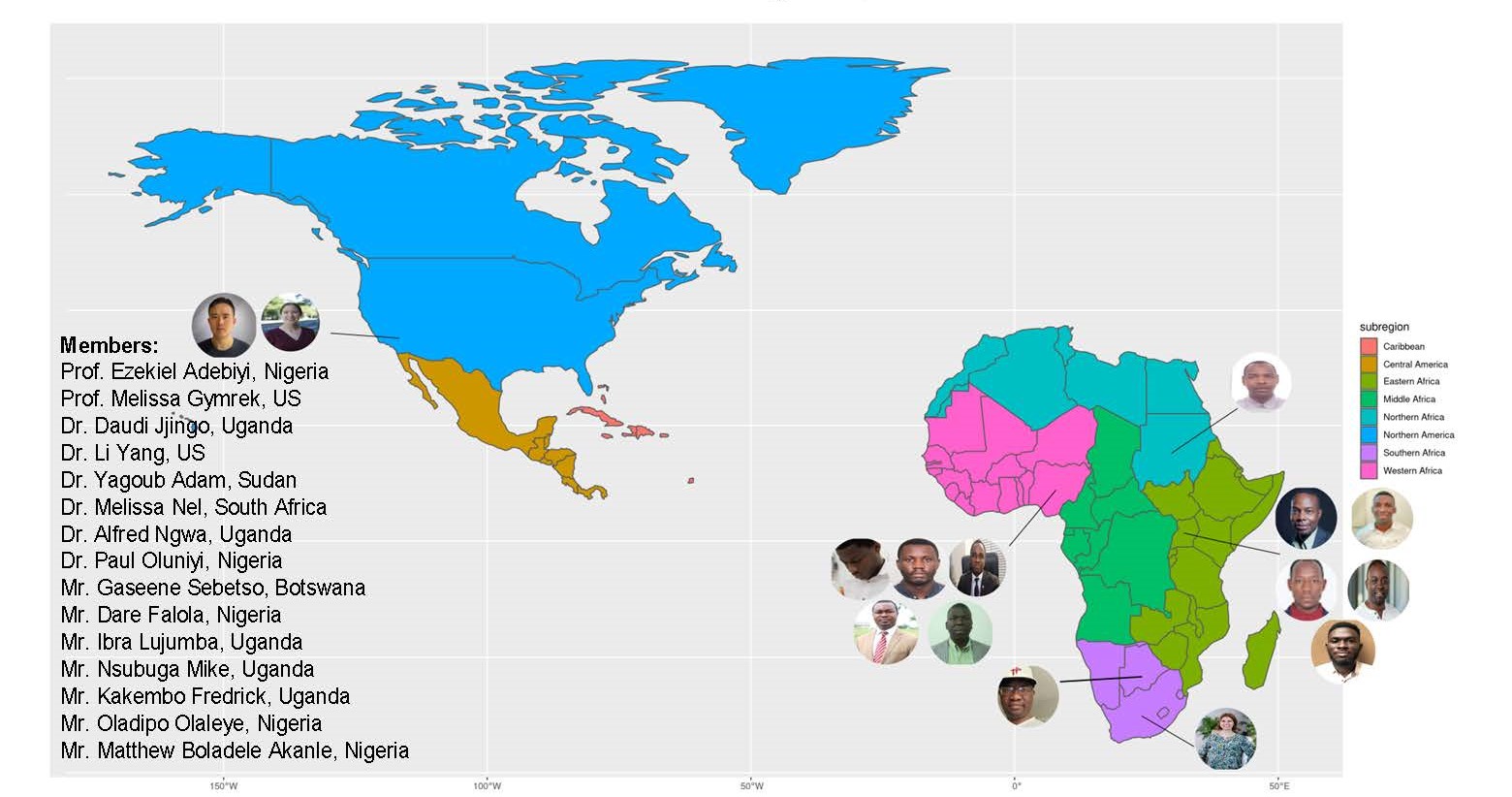
This project aims to characterize, on a genome-wide scale, novel tandem repeat expansions among Africa populations and how such expansions and other variants contribute to African phenotypes. This is a multi-site collaboration between personnel from Covenant University, Nigeria; University of Cape Town (UCT), South Africa; Makerere University, Kampala; University of California, San Diego, USA; Baylor College of Medicine, USA; Wellcome Genome Campus, UK and University of Witwatersrand, South Africa. Details on the people involved in the project can be found in Figure 1 below.
The research kicked off with the transfer of over 25TB of whole genome sequencing (WGS) data (H3A-Baylor and TrypanoGEN) from UCT to the high performance computing (HPC) facility at the African Center of Excellence (ACE) in Bioinformatics and Data Intensive Sciences at Makerere University, where the team is performing the analyses, in addition to using the HPC facility at UCT. To start, the biggest challenge was how to transfer the 25TB of data to Makerere University HPC. Therefore, to get the data transferred successfully, we employed three different methods, listed in the order that we were able to sort them for use, namely 1) rsync Linux utility, 2) shipping the data in external hard drive and 3) Globus transfer.
The Globus tool was our first option, as Globus is considered to be a fast and reliable data transfer tool. However, the enormous challenge in using Globus is its technical requirements and setting up procedure. To overcome this challenge, system administrators from Covenant University Bioinformatics Research (CUBRe) and the African Center of Excellence (ACE) in Bioinformatics and Data intensive Sciences, Makerere University, met couple of times to debugged issues arising, after the system administrators team from Makerere University did the first installation. The following are key resultants that helped to finalize the Globus installation for our use: 1) It is advisable to install Globus in a separate computer node because it uses specific setting, including firewall setting, network setting and system setting (the first installation was done on a computer node with other applications. Hence, the installation was not functional). 2) Upon Globus installation, endpoint ID and endpoint domain need to be created (and checked to be opened, preferably from another site different from the one with the Globus installation of interest) on the machine where Globus has been installed using this command: globus-connect-server endpoint setup. 3) To complete the setting up, another computing node should be added to host the targeted data. Adding a data transfer node to the Globus endpoint can be done using the following command: globus-connect-server node setup. However, the latter step needs extra firewall and network setting by applying particular network ports (these ports, preferably need to be checked, to be opened (for data transfer) from another site). 4) The data should be secured by creating a user group for those having access right to the data sets.
While debugging the Globus installation problems, we tried to use rsync Linux utility in parallel for the data transfer. Nevertheless, we stopped the rsync transfer once we resolved the issues with Globus. We found that Globus transfer is about 6 times faster compared to the original rsync transfer. At the initial stage of starting the Global transfer, we discovered that the transfer was terribly affected by the available internet speed. Therefore, we estimated that the transfer will be completed in 3 months. At this stage, we decided to explore the option of couriering external hard drive (the time to copy the data into the drive was about 3 weeks) from UCT to Makerere University. While working out this option, we upgraded the speed to 4.5MBps. Although such speed is still slow, we succeeded in using Globus to move the data across the two servers, bumping up of the bandwidth speeds from the Ugandan NREN made the complete transfer possible after close to a month.
Based on the above experience, Globus tool is the still, the tool of choice, for transferring huge data sets across different computing sites, even in Africa.
